Claude Pasquier
Researcher in Computer Science / Computational Biology
Université Côte d'Azur
CNRS
I3S laboratory
Biography
Claude Pasquier is researcher at French National Center for Scientific Research (CNRS).
He received a Ph.D. degree in Computer Science from the University of Nice - Sophia Antipolis (now Université Côte d’Azur), France, in 1994. During his thesis, he explored the use of software engineering paradigms in the field of structured document manipulation. He defined both a frame-based language for representing document models, and a system of context-aware document generation and configuration (cf. Prototype-based programming.
Subsequently, he was a postdoctoral researcher at the Biophysics and Bioinformatics Laboratory of the University of Athens, Greece, where he conducted research on protein structure prediction.
He held positions at the National Institute for Research in Digital Science and Technology (INRIA) and with Schlumberger, Smart Cards & Terminals division (now Gemalto) where he worked on generative programming.
Since 2002 when he joined CNRS, he successively worked at Villefranche Oceanographic Laboratory (LOV), the Institute of Biology Valrose (iBV) and New Caledonia Institute of Exact and Applied Sciences (ISEA) where he addressed topics as diverse as semantic data integration, omics data mining and attributed graph mining.
Currently at I3S laboratory, he is conducting research focused on complex network mining that combines computer science and systems biology.
Interests
- Data Mining
- Machine Learning
- Computational Biology
Education
HDR in Computer science, 2018
Université Côte d'Azur
PhD in Computer science, 1994
University of Nice - Sophia Antipolis
Projects
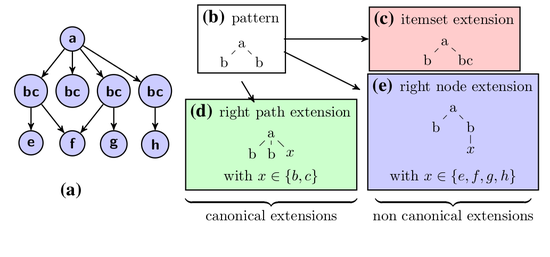
Attributed Graph Mining
Combining itemset mining and structural mining to reveal patterns in attributed graphs.
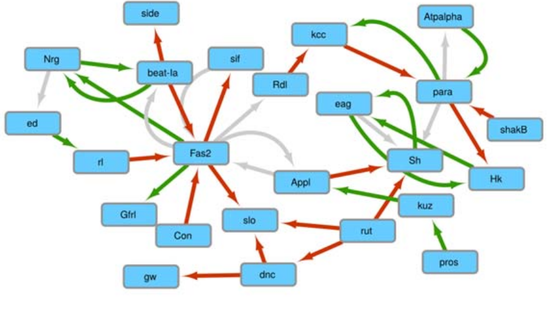
Complex Network Mining
Multidimensional networks mining in different fields of application.
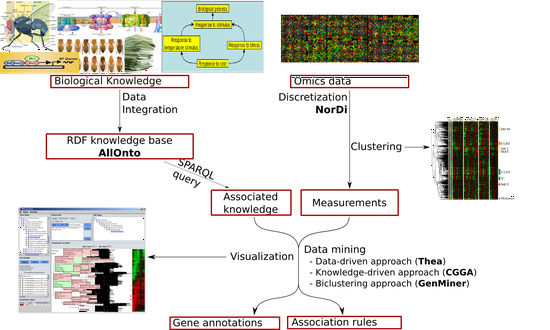
Omics Data Mining
Semantic integration and analysis of biological data.
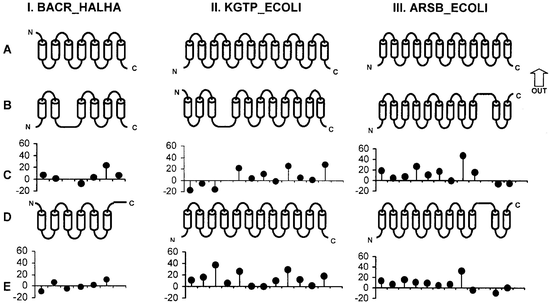
Protein Structure Prediction
Prediction of localization and topology of transmembrane alpha-helices in proteins.
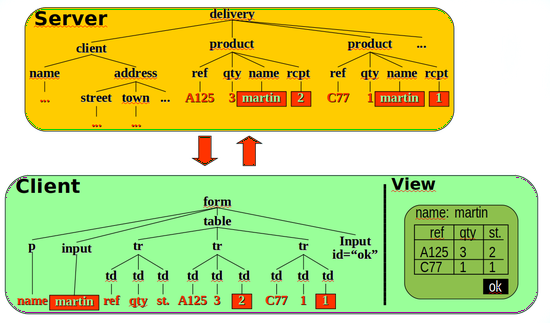
Generative Programming
Automatic generation of software tools from specifications.
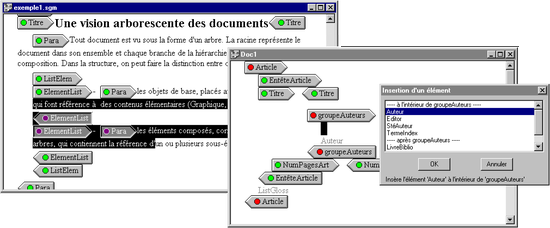
Prototype-based Programming
Dynamic document sharing based on delegation and cloning.
Recent Publications
Softwares
AMINE
Active Module Identification through Network Embedding.
AADAGE
Mining frequent patterns in attributed graphs.
AllOnto
Triple store with OWL query answering.
Bible
Management of Context-Controlled Documents.
CGGA
Extraction of bi-clusters of genes.
CoPreThi
Prediction of alpha-helices in proteins.
Dam-Bio
Protein sequence analysis on the Web.
DB-NTMR
Database of proteins' non transmembrane regions.
DB-TMR
Database of proteins' transmembrane regions.
FT
Periodicity analysis in molecular sequences.
GeniaJ
Java implementation of the Genia tagger.
GenMiner
Mining association rule from gene expression.
IMIT
Mining frequent patterns in attributed trees.
KeTuK
Mapping XML documents with a set of Java Beans.
KeyStract
Keywords extraction from scientific papers.
MiRAI
Prediction of microRNA-disease associations.
NorDi
Discretizing gene expression data.
OrienTM
Topology prediction of transmembrane proteins.
PRED-CLASS
Classification of proteins with a neural network.
PRED-TMR
Prediction of proteins' transmembrane regions.
PRED-TMR2
Identification of transmembrane proteins.
THEA
Automatic annotation of genes.
THEA-INTERACT
Analysis of gene interaction network.
THEA-ONLINE
Semantic data integration.